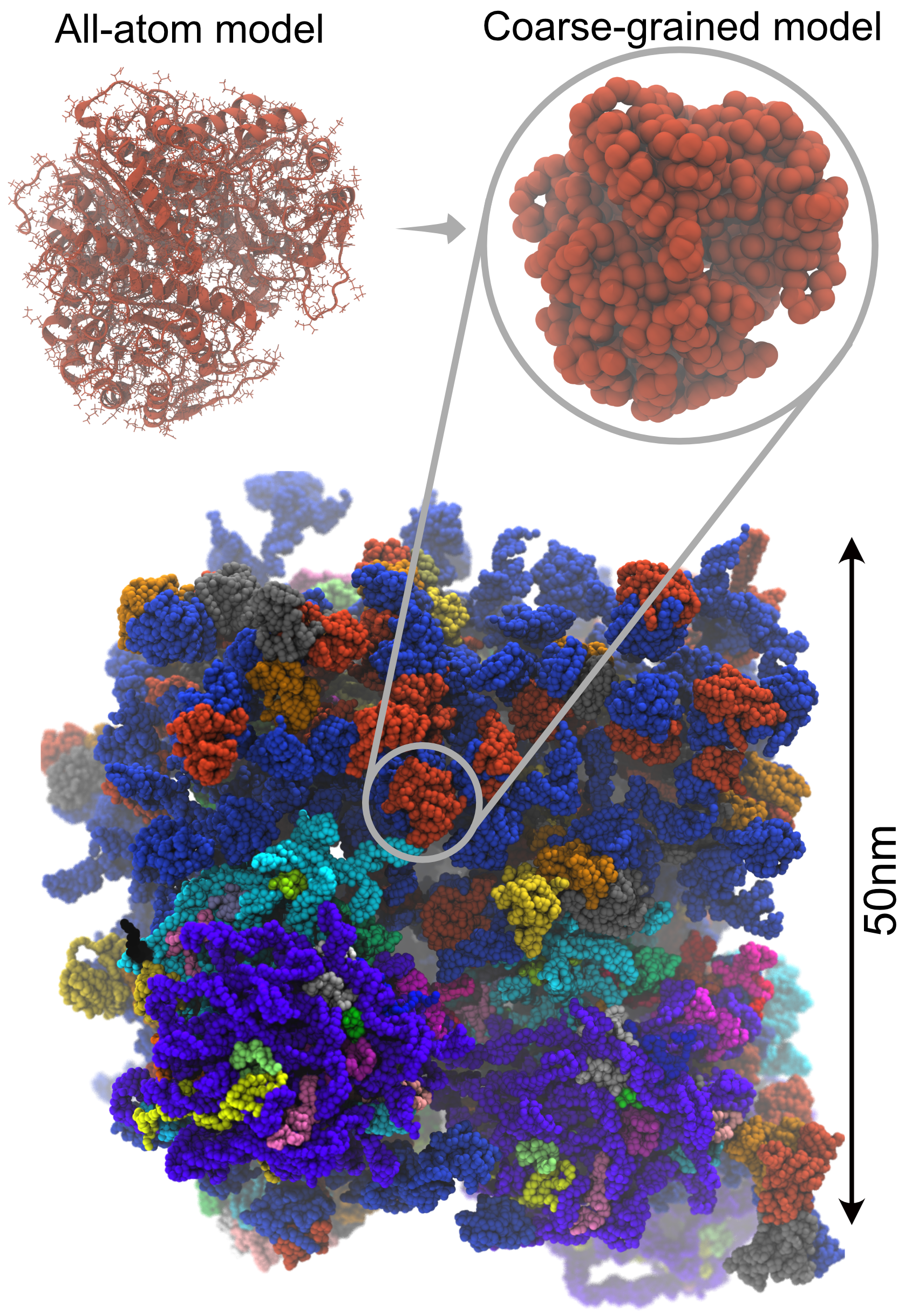
Inside the cell, macromolecules such as proteins, nucleic acids, and lipids interact with each other. It is thought that molecules change their reactivity and stability by recognizing each other, and understanding the mechanism of such processes, which are essential for biological activity is a fundamental issue. However, studying environments which include numerous macromolecules of different types is very difficult due to the enormous amount of calculation required. We develop coarse-grained models which extend the time and spatial range observable by simulation. Specifically, we develop multi-resolution methods which integrate models of different resolutions including all-atom models and the quantum chemical method QM/MM, and implement them into GENESIS. Using these methods, we aim to perform cellular-scale simulations which include numerous molecule types, using the supercomputer Fugaku.
References
- “A singularity-free torsion angle potential for coarse-grained molecular dynamics simulations.”, Cheng Tan, Jaewoon Jung, Chigusa Kobayashi, and Yuji Sugita, J. Chem. Phys., 153, 044110 (2020).
- “Scaling Molecular Dynamics Beyond 100,000 Processor Cores for Large-Scale Biophysical Simulations”, Jaewoon Jung, Wataru Nishima, Marcus Daniels, Gavin Bascom, Chigusa Kobayashi, Adetokunbo Adedoyin, Michael Wall, Anna Lappala, Dominic Phillips, William Fischer, Chang-Shung Tung, Tamar Schlick, Yuji Sugita, and Karissa Y. Sanbonmatsu, J. Comput. Chem., 40, 1919-1930 (2019).
- “Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm”, Isseki Yu, Takaharu Mori, Tadashi Ando, Ryuhei Harada, Jaewoon Jung, Yuji Sugita, and Michael Feig., eLife, 5, e19274 (2016).
- “Domain Motion Enhanced (DoME) Model for Efficient Conformational Sampling of Multidomain Proteins”, Chigusa Kobayashi, Yasuhiro Matsunaga, Ryotaro Koike, Motonori Ota, and Yuji Sugita, J. Phys. Chem. B, 119, 14584-14593 (2015).